When a new disease comes out of the woodwork, scientists across the world leap into action to try and find out what they can about it, hopefully providing new ways to help.
Researchers at the University of Minnesota (UM) have done just that – by investigating the structure of the 'spike' protein on the surface of SARS-CoV-2, the team hopes they've contributed to the groundwork for new drug design.
"In general, by learning what structural features of viral proteins are most important in establishing contact with human cells," explains UM biomedical researcher Fang Li, "we can design drugs that seek them out and block their activity - like jamming their radar."
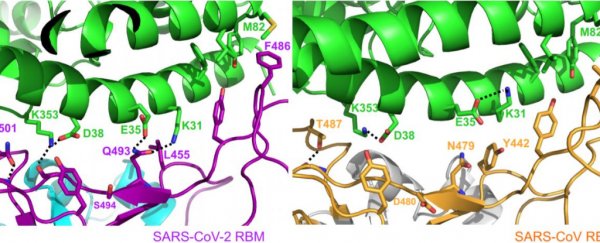
The team used X-ray crystallography to create a 3D model of what the spike protein looks like, and just how it binds to human cells. (You can see the finished product above.)
Although this doesn't look like the pictures of the coronavirus you're used to seeing, it's an incredibly helpful model for biologists. It allows them to visualise how small mutations in the protein create different folds and ridges, which then change the way the virus particle attaches to receptors in our own cells.
What the team found is that the SARS-CoV-2 strain of coronavirus has a few mutations that form a particularly compact 'ridge' in the spike protein.
This ridge is more compact than the one in the SARS virus, and this could be one of the reasons this new strain is so adept at infecting humans, causing COVID-19.
"The 3D structure shows that compared to the virus that caused the 2002-2003 SARS outbreak, the new coronavirus has evolved new strategies to bind to its human receptor, resulting in tighter binding," Li told The Guardian.
"The tight binding to the human receptor can help the virus infect human cells and spread among humans."
The team also looked at similar strains of the coronavirus in bats and pangolins, finding that the bat strain would have to go through a number of mutations to arrive at a spike shape that fits the human receptor well.
However, one particular strain of the pangolin virus had a better human receptor fit, giving a little more leverage to the hypothesis that pangolins were an intermediate host for the virus.
The team hopes that the new modelling will help other researchers develop drugs or vaccines for the virus.
"Our work can guide the development of monoclonal antibodies that would act like a drug to recognise and neutralise the receptor-binding part of the spike protein," Li said.
"Or, a part of the spike protein could become the basis of a vaccine."
But we need to be cautious at this stage. This sort of research is constantly evolving, and although the model is promising, the study only used small fragments of the virus spike - its binding domain - and so there is likely more information left to discover.
We're sure scientists all over the world are racing to uncover it, so we can all get through this together.
The research has been published in Nature.