The acronym CRISPR has become synonymous with editing DNA in recent years, taking center stage in the molecular geneticist's toolbox as a means of identifying genetic codes and then cutting into them with uncanny precision.
In its original function as a means of immunity in bacteria, the CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated endonuclease) system seeks out known genes of invading viruses and renders them dysfunctional.
Scientists from the University of Rochester and Cornell University, Ithaca, in the US and have discovered that the famous gene-editing tool does more in bacteria than just spot DNA for chopping up; it coordinates with other proteins to bulk up defenses against invading viruses as well.
Activating the funnel-shaped proteins – called Csx28 – scrambles the permeability of the bacteria's membrane, making it difficult for invading viral DNA to hijack the cell's machinery and replicate.
The finding, published in Science, is "unexpected and raises all kinds of new questions," says University of Rochester biochemist Mark Dumont, a collaborator on the study.
"While there is no immediate medical relevance or application, the ideas that boil up from this could be very powerful," Dumont says.
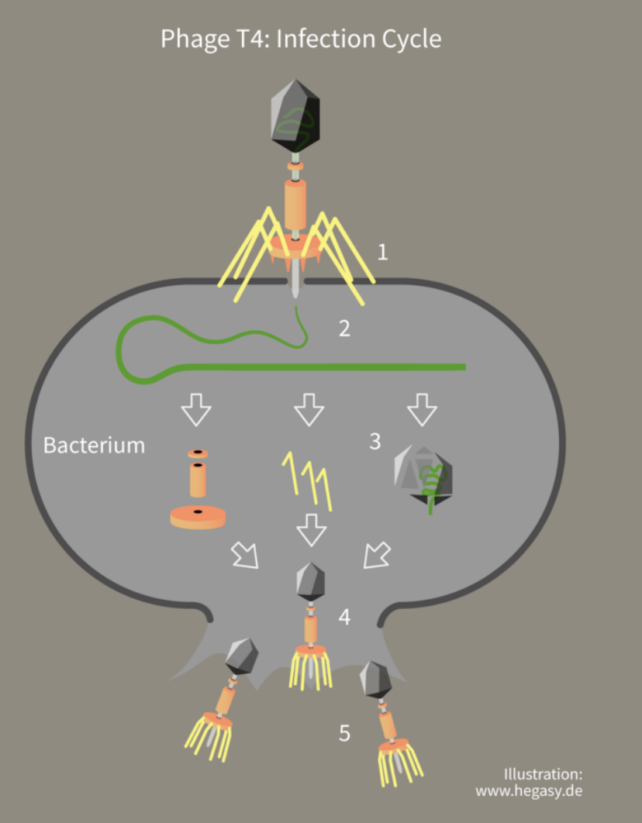
The study involved a series of experiments where Escherichia coli bacteria were infected with a virus that infects bacteria, or bacteriophage, called enterobacteria phage λ.
This phage latches onto the bacteria cell surface like a lunar module and injects its DNA into the cell to create copies of itself.
The E. coli fight back, using CRISPR to identify the threat by matching repetitive DNA sections from previously encountered phages, and then using an enzyme called Cas13b to snip the invading DNA into pieces.
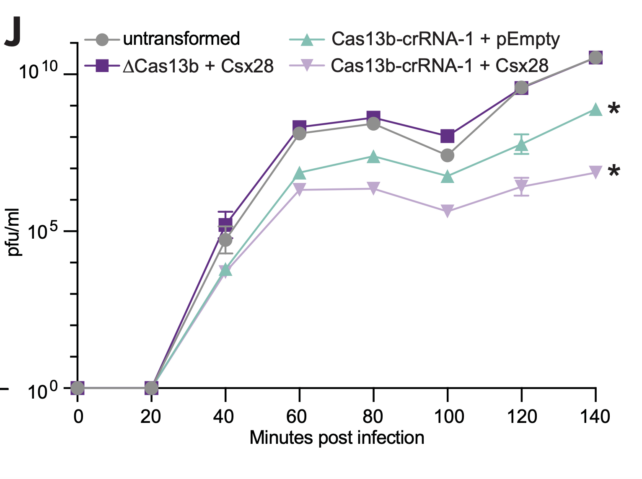
The researchers found that the virus replicated sluggishly when Csx28 was present inside the bacteria.
This protein only worked in conjunction with Cas13b, which suggested the two were coordinating with each other to disarm the virus.
When both Cas13b and Csx28 were present, the proportion of infected bacteria that released infectious viral particles decreased from around 19 percent to roughly 3 percent, and there was a significant reduction of phage numbers per milliliter. In other words, the virus wasn't able to replicate as much as it usually would.
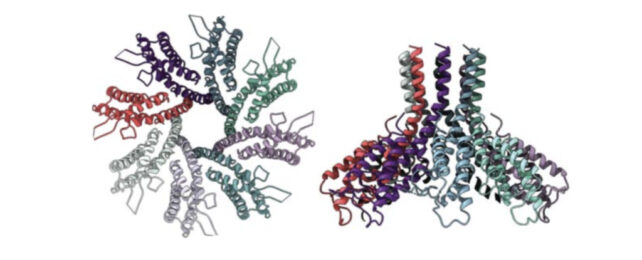
- The researchers examined the structure of the Csx28 protein using a technique called cryogenic electron microscopy and found that it resembled a funnel with a hole in the center.
This raised the possibility that the protein formed a membrane pore and was disrupting the metabolism of the cell to make it an inhospitable environment for the virus.
The researchers tested this hypothesis using a technique that makes cells fluorescent after they have lost their membrane potential, a small electrical charge caused by the difference in the concentration of ions inside and outside the cell.
They found that the two proteins together caused the membrane to depolarize, sending in a rush of charged atoms that radically altered the cell's internal environment. After 90 minutes, 40 percent of the bacterial population was depolarized in this fashion.
"When you read this paper you think to yourself…'What?' This is such a weird mechanism," says University of Rochester molecular biologist John Lueck, who wasn't involved in the research.
"It is really impressive that the team identified this pore-like protein that doesn't resemble anything else we've seen before," he says. "This is exciting because in science, when you scratch the surface, you often find that there is an entirely new world behind it."
This paper was published in Science.