Once a leading cause of death for children across the western world, scarlet fever was nearly eradicated thanks to 20th century medicine. But fresh outbreaks in the UK and North East Asia over recent years suggest we've still got a long way to go.
Just why we're experiencing a resurgence of the deadly pathogen is a mystery. A new study has uncovered clues in the genome of one of the bacterial strains responsible, showing just how complex the family tree of infectious diseases can be.
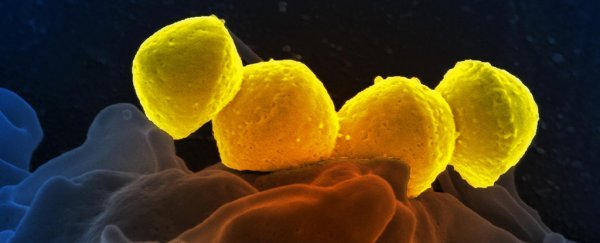
The species behind the illness is group A strep, or Streptococcus pyogenes; a ball-shaped microbe that can churn out toxic compounds called superantigens, capable of wreaking havoc inside the body. Especially in children.
The results can be as mild as an uncomfortable case of pharyngitis or a bad rash, or as severe as a toxic shock that causes organs to fail.
With the advent of antibiotics, outbreaks could easily be managed before they got out of hand. By the 1940s, the disease was well on the way out.
That all looks to be changing.
"After 2011, the global reach of the pandemic became evident with reports of a second outbreak in the UK, beginning in 2014, and we've now discovered outbreak isolates here in Australia," says University of Queensland molecular biologist Stephan Brouwer.
"This global re-emergence of scarlet fever has caused a more than five-fold increase in disease rate and more than 600,000 cases around the world."
Leading an international team of researchers in a study on group A strep genes, Brouwer has been able to characterise a variety of superantigens produced by one particular strain from North East Asia.
Among them was a kind of superantigen that appears to give the bacterial invaders a clever new way to gain access to the insides of the host's cells, one never seen before among bacteria.
Its novelty implies that these outbreaks aren't descended from the same strains of bacteria that have rippled through communities in centuries past. Rather, they're closely related populations of group A strep that learned a new trick or two on their own.
One way similar organisms can evolve the same characteristics – such as advanced virulence – is for natural selection to independently fine-tune shared genes in the same way.
But other studies have already suggested this strain of bacterium received a helping hand in the form of an infection of their own, one from a type of virus called a phage.
"The toxins would have been transferred into the bacterium when it was infected by viruses that carried the toxin genes," says bioscientist Mark Walker, also from the University of Queensland.
"We've shown that these acquired toxins allow Streptococcus pyogenes to better colonise its host, which likely allows it to out-compete other strains."
In a process known as horizontal gene transfer, a gene that evolved in one microbe can be incorporated into a virus's genome and edited into a new host's DNA, creating a kind of clone of the original.
Though hardly limited to bacteria, it is a quick and handy way for single-celled microbes to adapt. Such stolen genes can provide pathogens with new ways to gain entry to host tissues, or resist the chemical warfare that would otherwise keep them at bay.
In this case, it has helped a less serious strain of bacteria to develop a weapon that makes it as concerning as its vanquished cousin.
To double check the acquired superantigen's importance, the researchers used genetic editing to disable their coding. As a result, the strains lost their knack for colonising the animal models used to test the bacteria's virulence.
For now, our management of an even bigger threat seems to be containing the most recent scarlet fever outbreaks. Spread through aerosols much like SARS-CoV-2, group A strep is unlikely to become an epidemic under current restrictions.
"But when social distancing eventually is relaxed, scarlet fever is likely to come back," says Walker.
"Just like COVID-19, ultimately a vaccine will be critical for eradicating scarlet fever – one of history's most pervasive and deadly childhood diseases."
This research was published in Nature Communications.